import anon
anon.conf.config.update('jax_disable_jit', True)
anon.conf.config.update("jax_enable_x64", True)
import anon.atom as anp
import elle.sections
import elle.beam2d
import anabel as em
geom_template = elle.beam2d.transform_no2(elle.beam2d.geom_no2)
beam_template = elle.beam2d.resp_no1An Assembler is an object with an .assemble() and optionally a .compose() method
# Create a model Assembler
model = em.SkeletalModel(ndm=2,ndf=3)
# Define problem parameters
ColumnE = model.param("ColumnE")
GirderE = model.param("GirderE")
P1 = model.param("P1")
# Define model components
girder_section = em.aisc.load("w33x130")
column_section = em.aisc.load("w14x159")
basic_girder = beam_template(**girder_section, E=GirderE)
basic_column = beam_template(**column_section, E=ColumnE)
girder = geom_template(basic_girder)
column = geom_template(basic_column)WARNING:absl:No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)# Set up nodes
ft = 12
B, H = 30.*ft, 13.*ft
model.node("1", 0., 0.)
model.node("2", 0., H )
model.node("3", B/2, H )
model.node("4", B , H )
model.node("5", B , 0.)
model.beam("a", "1", "2", **column_section, elem=column)
model.beam("b", "2", "3", **girder_section, elem=girder)
model.beam("c", "3", "4", **girder_section, elem=girder)
model.beam("d", "4", "5", **column_section, elem=column)
model.boun("1", [1,1,1])
model.boun("5", [1,1,1])
model.load("2", P1, dof="x")
model.load("2", -2e3, dof="y")
model.load("4", -2e3, dof="y")
em.plot_skeletal(model);
f = model.compose()f(3600.,3600.,2e3)/home/claudio/elle/numeric/elle/numeric/inverse.py:111: UserWarning: Function inversion failed to converge.
warnings.warn("Function inversion failed to converge.")
DeviceArray([[ 0.28355172],
[ 171.74283748],
[ 0.84689206],
[-202.66515683],
[-149.92389898],
[ 5.04060219],
[ -14.82161937],
[-148.43045677],
[ 5.1556104 ]], dtype=float64)fig, ax = em.plot_displ(model, f(29e6,29e6,20e3),scale=1e3)/home/claudio/elle/numeric/elle/numeric/inverse.py:111: UserWarning: Function inversion failed to converge.
warnings.warn("Function inversion failed to converge.")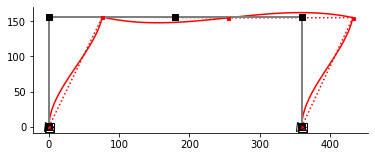
df = anon.diff.jacfwd(f, None, (0,1),squeeze=False)df(29e6,29e6,20e3)dx: Traced<ConcreteArray([[20000.]
[-2000.]
[ 0.]
[ 0.]
[ 0.]
[ 0.]
[ 0.]
[-2000.]
[ 0.]])>with<JVPTrace(level=2/0)>
with primal = DeviceArray([[20000.],
[-2000.],
[ 0.],
[ 0.],
[ 0.],
[ 0.],
[ 0.],
[-2000.],
[ 0.]], dtype=float64)
tangent = Traced<ShapedArray(float64[9,1])>with<BatchTrace(level=1/0)>
with val = DeviceArray([[[-0.],
[-0.]],
[[-0.],
[-0.]],
[[-0.],
[-0.]],
[[-0.],
[-0.]],
[[-0.],
[-0.]],
[[-0.],
[-0.]],
[[-0.],
[-0.]],
[[-0.],
[-0.]],
[[-0.],
[-0.]]], dtype=float64)
batch_dim = 1
/home/claudio/elle/numeric/elle/numeric/inverse.py:112: UserWarning: Function inversion failed to converge.
warnings.warn("Function inversion failed to converge.")
(DeviceArray([[-2.04476205e-09],
[-1.10382234e-11],
[ 7.00882848e-13],
[-2.04415680e-09],
[-5.35148746e-12],
[-2.71208496e-13],
[-2.04355123e-09],
[ 2.30895445e-11],
[ 9.53551267e-13]], dtype=float64),
DeviceArray([[-5.79214813e-10],
[ 9.60734398e-13],
[ 7.15495715e-12],
[-5.24811184e-10],
[ 3.78153916e-11],
[-3.37652007e-12],
[-4.70407107e-10],
[-1.95779547e-12],
[ 6.30254346e-12]], dtype=float64))